Ground-truth recordings for validation of spike sorting algorithms
This dataset is composed of simultaneous loose patch recordings of Ganglion Cells in mice retina, combined with dense extra-cellular recordings (252 channels). The details of the dataset can be found here https://elifesciences.org/articles/34518
Probe layout
The probe layout can be found as mea_256.prb. This is a 16x16 Multi Electrode Array with 30um spacing. Only 252 channels are extra-cellular signals, and the 4 corners are devoted to triggers/sync/juxta.
Struture of the data
In this dataset, you will find several individual recordings, at max 5min long each (but please do not hesitate to contact us if interested by longer recordings). The extra-cellular data are saved as 16bits unsigned integer, with a variable offset at the beginning of the file. The value of this offset is given, for every datafile, in the additional text file (padding value (see following for more details)). The files have already been filtered with a Butterworth filter of order 3 with a cut-off frequency at 100Hz
Structure of a given dataset
Please read carefully the following to understand how to load and perform spike sorting with the data. In every .tar.gz file, you will find:
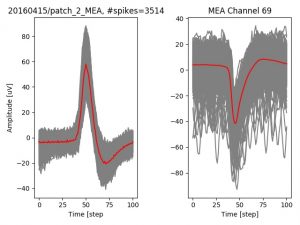
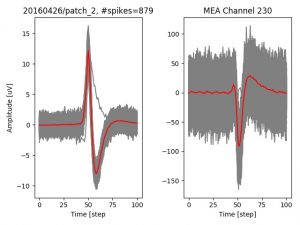
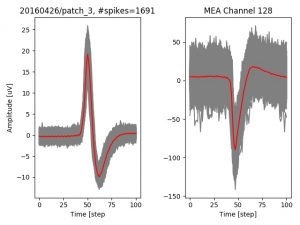
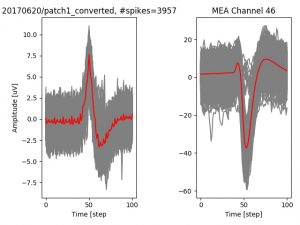
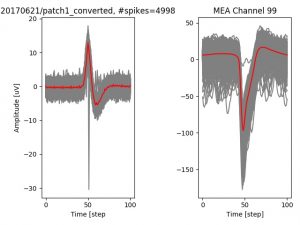
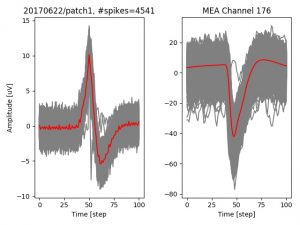
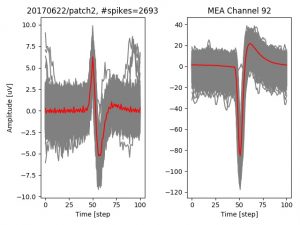
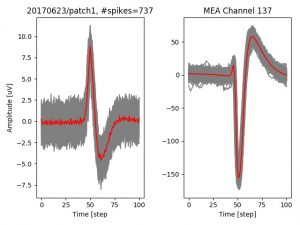
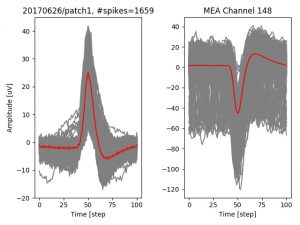
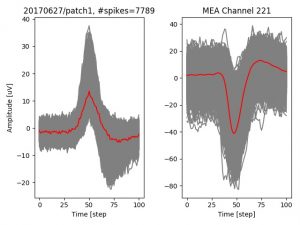
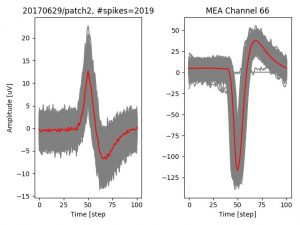
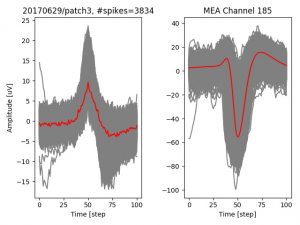
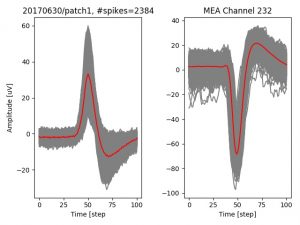
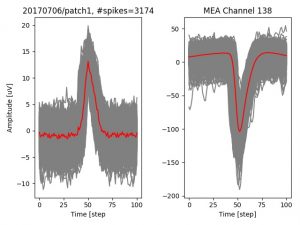
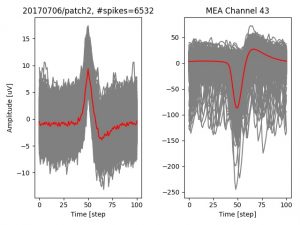
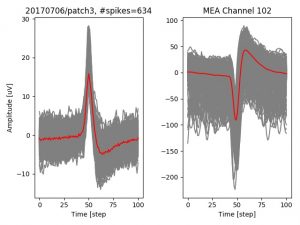
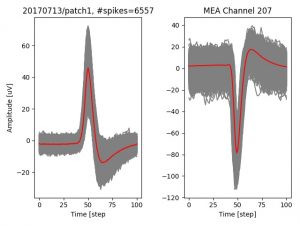
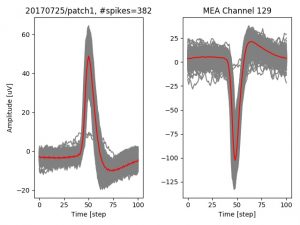
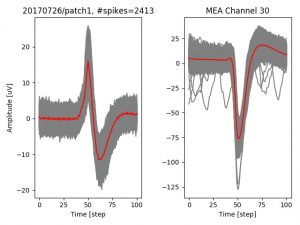
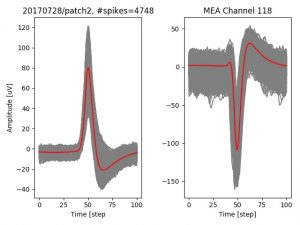
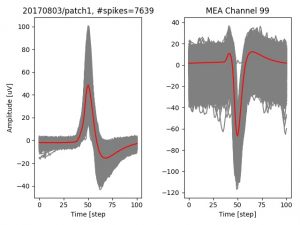
- a jpg image, displaying a small chunk of the juxta-cellular signal (top left), with detected peaks and threshold. The extra-cellular spike triggered waveform, across all channels, for the juxta-spike times (top right). In the bottom, you can see the juxta-cellular spikes, for all the detected triggers (left), and on the right the voltage on the channel where the Spike Triggered Average of the extra-cellular waveform is peaking the most.
- a file .juxta.raw, as float32, with the juxta-cellular trace at 20kHz, no data offset
- a file .raw, as uint16, with the extra-cellular signals recorded for 256 channels at a sampling rate of 20kHZ. In fact, only 252 channels are extra-cellular signals, the 4 corners of the arrays are devoted to juxta-cellular and sync signals (see probe layout mea_256.prb)
- a file .triggers.npy containing the spike times of the juxta-cellular spikes, detected using a threshold of k.MAD. The exact value of k can vary on a per dataset basis, and is written in the .txt file (threshold)
- a .txt file describing some information for a given dataset, such as the threshold value used to detect the spikes, the channel in the raw file where the juxta-cellular signal is located, the minimal value of the peak for the STA (and on which channel it is located), and the header size to read the raw data
- a .params file, if you want to analyze the data with SpyKING CIRCUS
-
20160415
-
20160426
-
20170620
-
20170621
-
20170622
-
20170623
-
20170626
-
20170627
-
20170629
-
20170630
-
20170706
-
20170713
-
20170725
-
20170726
-
20170728
-
20170803